Single cell technology development
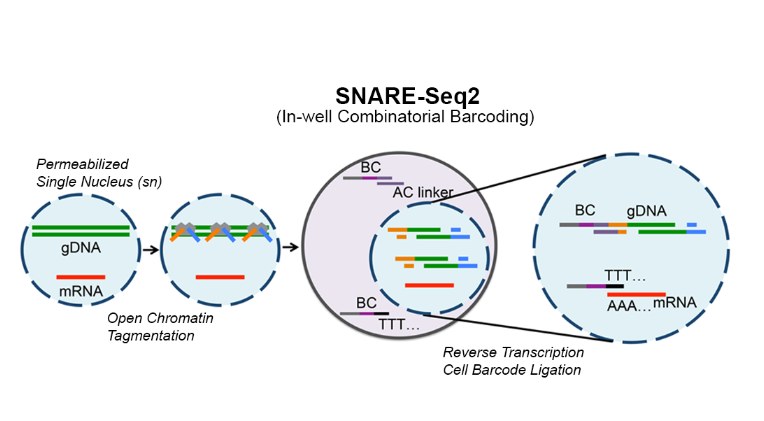
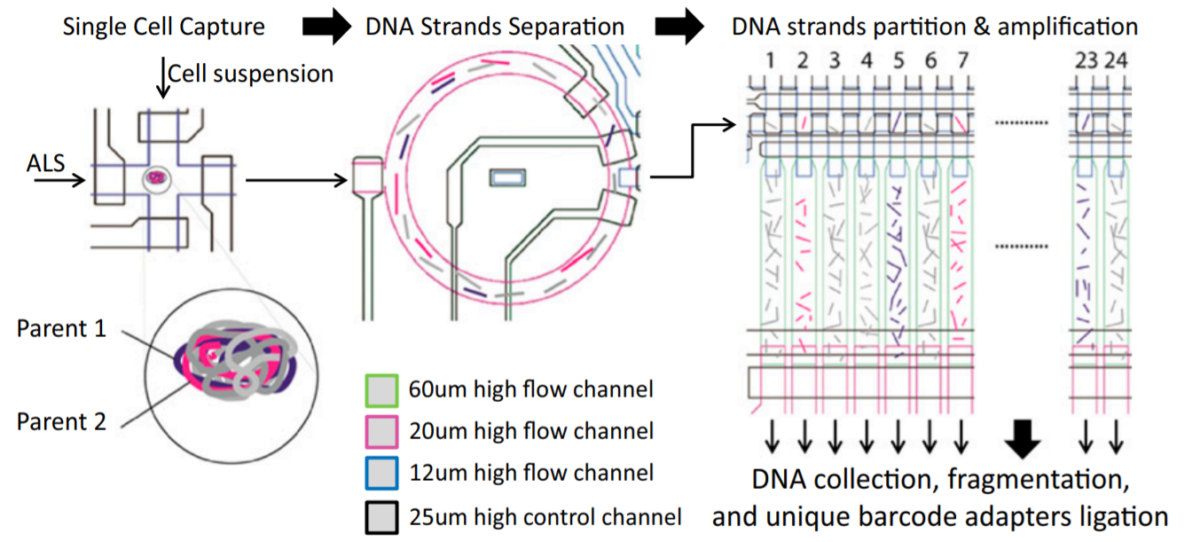
Human cells take on a variety of cell states depending on their function (neurons, gut epithelium, etc) as well as other factors such as the presence of disease, chemical or genetic perturbations, the cellular microenvironment, etc. In order to capture cell state and be able to explain for example, the differences between injured kidney cells and healthy kidney cells, we need to be able to capture cell states in single cells. And because there are so many different cells with different functions and an almost infinite number of factors that can shift those cell states, we need to measure cell states across many cells. Thus, our lab develops high-throughput technologies to capture cell state by measuring biomolecules such as RNA, chromatin accessibility, methylation, or genomic mutations. We have also developed technologies that can capture multiple types of biomolecules in the same cell. For example, our SnareSeq and SnareSeq2 methods can capture RNA and chromatin accessibility in the same cell, enabling us to link gene expression with the activity of noncoding regulatory elements like enhancers and promoters.
Mapping human cell types using single cell technologies
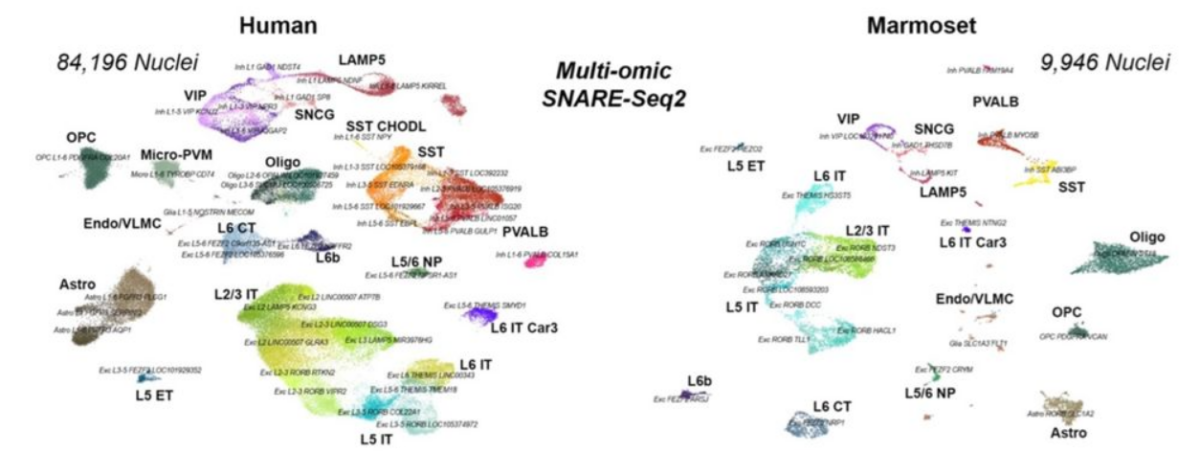
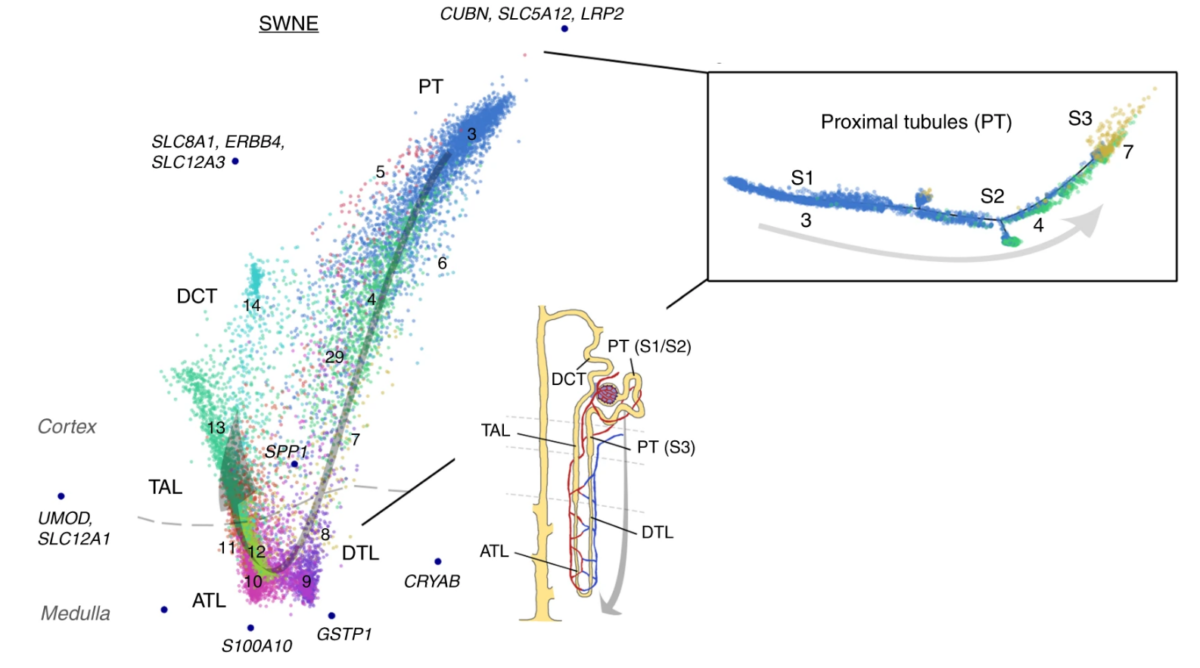
We apply our single cell and spatial 'omics technologies towards generating maps of human cell types across the brain, kidney, lung, heart, and various pediatric cancers. These technologies generate large amounts of data that we then analyze and interpret using machine learning tools such as dimensional reduction, clustering, and classification. Our goal is to better understand cell state and how cells behave in both healthy and diseased human cells across all of these different cell types.
Read more about our human cell type maps here.
