Single cell technology development
High throughput methods for capturing RNA, chromatin accessibility, methylation, and genomic mutations in single cells
Single cell multi-omics
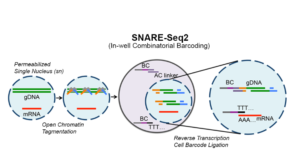
Being able to measure multiple types of biomolecules in the same cell enables us to better understand cell state as well as how these different types of molecules (such as RNA and protein) interact. We developed SnareSeq, a droplet based method for measuring both gene expression (via RNA-seq) and chromatin accessibility (via ATAC-seq) in the same cells. Linking these two modalities enables us to better understand gene regulation by allowing us to measure gene expression alongside the accessibility of regulatory elements like enhancers or promoters. We also developed SnareSeq2, which uses a combinatorial indexing platform, enabling us to profile the gene expression and chromatin accessibility of up to hundreds of thousands of cells in a single experiment.
Spatial transcriptomics
Cells are organized into higher level structures like tissues and organs, where the spatial organization of cells is critical to the ability of these tissues and organs to function. Thus, we need to be able to measure cell state while retaining the spatial organization of the cells. We are developing Dartfish, a method for profiling the gene expression across a tissue section, which enables us to understand how different cell types are spatially organized.
Single cell chromatin accessibility sequencing
Profiling the chromatin accessibility landscape of the genome enables us to identify regions of the genome that are active or inactive in a given cell. These regions oftentimes enhancers or promoters, and by profiling their activity we can better understand the gene regulatory landscape of cells. Additionally, certain diseases such as cancer will often result in disordered chromatin structure. We developed a method, THS-seq, that enables us to better capture chromatin accessibility profiles as compared with ATAC-seq. We also used a combinatorial indexing platform to develop single cell THS-seq, enabling us to profile the chromatin accessibility of tens of thousands of cells in a single experiment.
Functional genomics in multi-lineage organoids using CRISPR-Cas9 coupled with single cell RNA-seq readouts
One of the most robust ways to assess the function of a gene is to disable the gene via knockouts in some model system (i.e. cell lines, organoids) and measure the resulting changes in cell state using single cell RNA-seq. However, many genes have vastly different functions depending on the cellular context and existing human model systems such as cell lines and organoids can only capture a limited number of cell lineages. For example brain organoids can only model brain cell types. We established the teratoma as a multi-lineage model system that contains most major human cell lineages, and demonstrated that we could use teratomas to assess gene function across cell lineages using CRISPR-Cas9 screens.
Single nucleus RNA sequencing
For certain human organs like the human brain, it can be extremely difficult to isolate whole cells for single cell sequencing. Additionally, it is oftentimes necessary to freeze down tissue sections from donors and run single cell sequencing at a later date or at a different institution. In these cases, sequencing single nuclei instead of single cells can result in much higher data quality. Thus, we developed a method for single nucleus RNA sequencing by adapting the Drop-seq platform and applied this method to sequence human brain cells and kidney cells.
We previously built a method using the C1 platform and Smart-seq to generate a single nucleus RNA sequencing dataset from the human brain with full length mRNA profiling.
Visualizing single cell datasets
Visualizations of single cell datasets using non-linear dimensional reduction methods like t-Stochastic Neighbor Embedding (t-SNE) or Uniform manifold Approximation and Projection (UMAP) is critical to interpreting these datasets. We developed a non-linear dimensional reduction method, Similarity Weighted Nonnegative Embedding (SWNE) that captures the high dimensional structure of single cell datasets while allowing for the embedding of biological landmarks like marker genes or co-expressed gene modules.
Single cell DNA sequencing
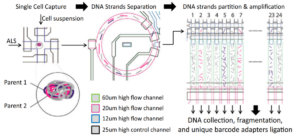
Genomic mosaicism is when some cells in your body have a slightly different genome than other cells. Oftentimes this occurs during diseases like cancer, where tumor cells will accumulate mutations since the cellular mechanisms preventing new mutations are broken. Mosaicism has also been shown to occur in neurodegenerative diseases like Alzheimer's Disease, and being able to profile the genome of individual cells can help us understand the mutations driving some of these diseases. We developed MIDAS, a method for uniform whole genome sequencing in single cells using microwells. However, many key mutations are known as point mutations in that they only change a single DNA nucleotide. In order to detect these more subtle mutations, we developed SISSORS, a microfluidic platform that can sequence the single strands from both homologous chromosomes, giving us multiple redundant sources of genomic information in each cell. Thus, SISSORS allows us to accurately identify point mutations in individual cells.

Bisulfite Padlock Probes (BSPP)
(Published in Nature Methods)

Allele-Specific Methylation (ASM)
(Published in Genome Research)